Across kingdom biased CYP-mediated metabolism via small-molecule ligands docking on P450 oxidoreductase
Abstract
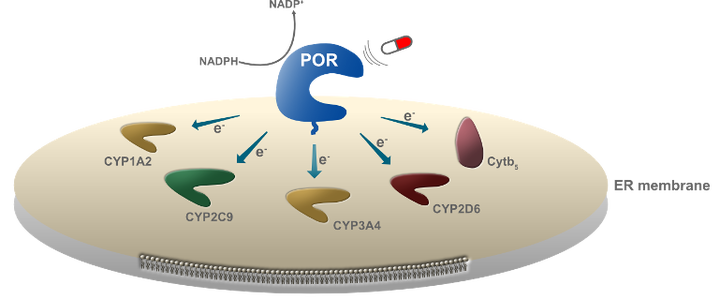
Metabolic control is mediated by the dynamic assemblies and function of multiple redox enzymes. A key element in these assemblies, the P450 oxidoreductase (POR), donates electrons and selectively activates numerous (>50 in humans and >300 in plants) cytochromes P450 (CYPs) controlling metabolism of drugs, steroids and xenobiotics in humans and natural product biosynthesis in plants. The mechanisms underlying POR-mediated CYP metabolism remain poorly understood and to date no ligand binding has been described to regulate the specificity of POR. Here, using a combination of computational modeling and functional assays, we identified ligands that dock on POR and bias its specificity towards CYP redox partners. Single molecule FRET studies revealed ligand docking to alter POR conformational sampling, which resulted in biased activation of metabolic cascades in whole cell assays. We propose the model of biased metabolism, a mechanism akin to biased signaling of GPCRs, where ligand docking on POR stabilizes different conformational states that are linked to distinct metabolic outcomes. Biased metabolism may allow designing pathway-specific therapeutics or personalized food suppressing undesired, disease related, metabolic pathways.